TiClNet
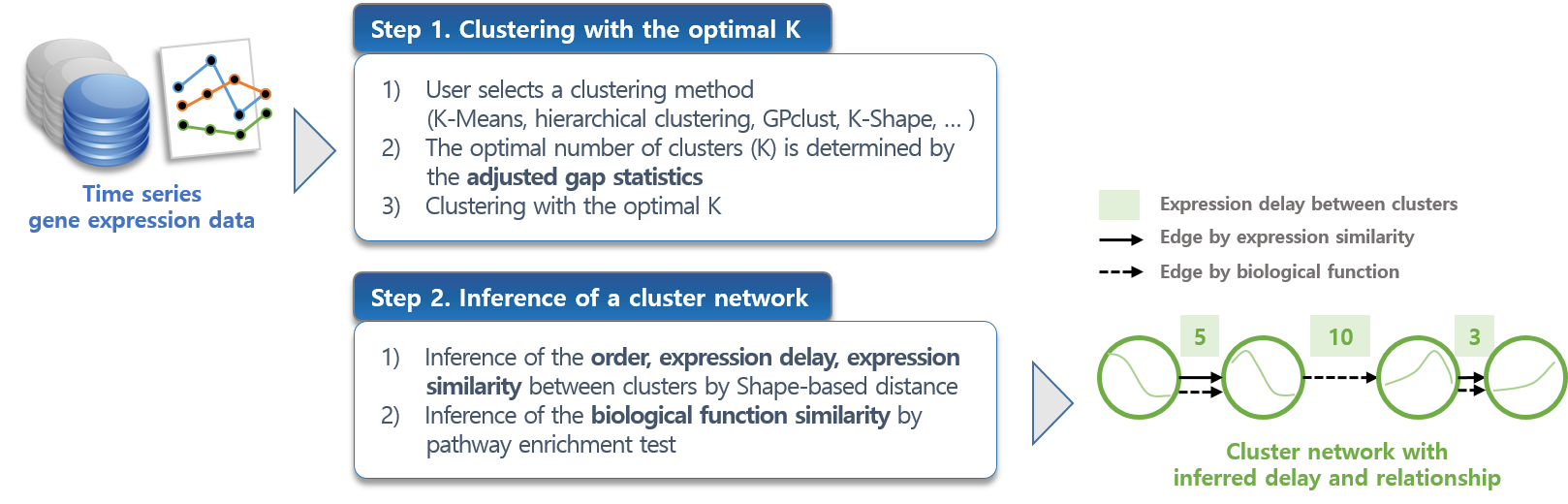
|
Time series-specific Clustering and Network inference (TiClNet) aims to solve the problems that occur at each step of clustering time series gene expression data as follows: |
Pathway Logic analysis on breast cancer
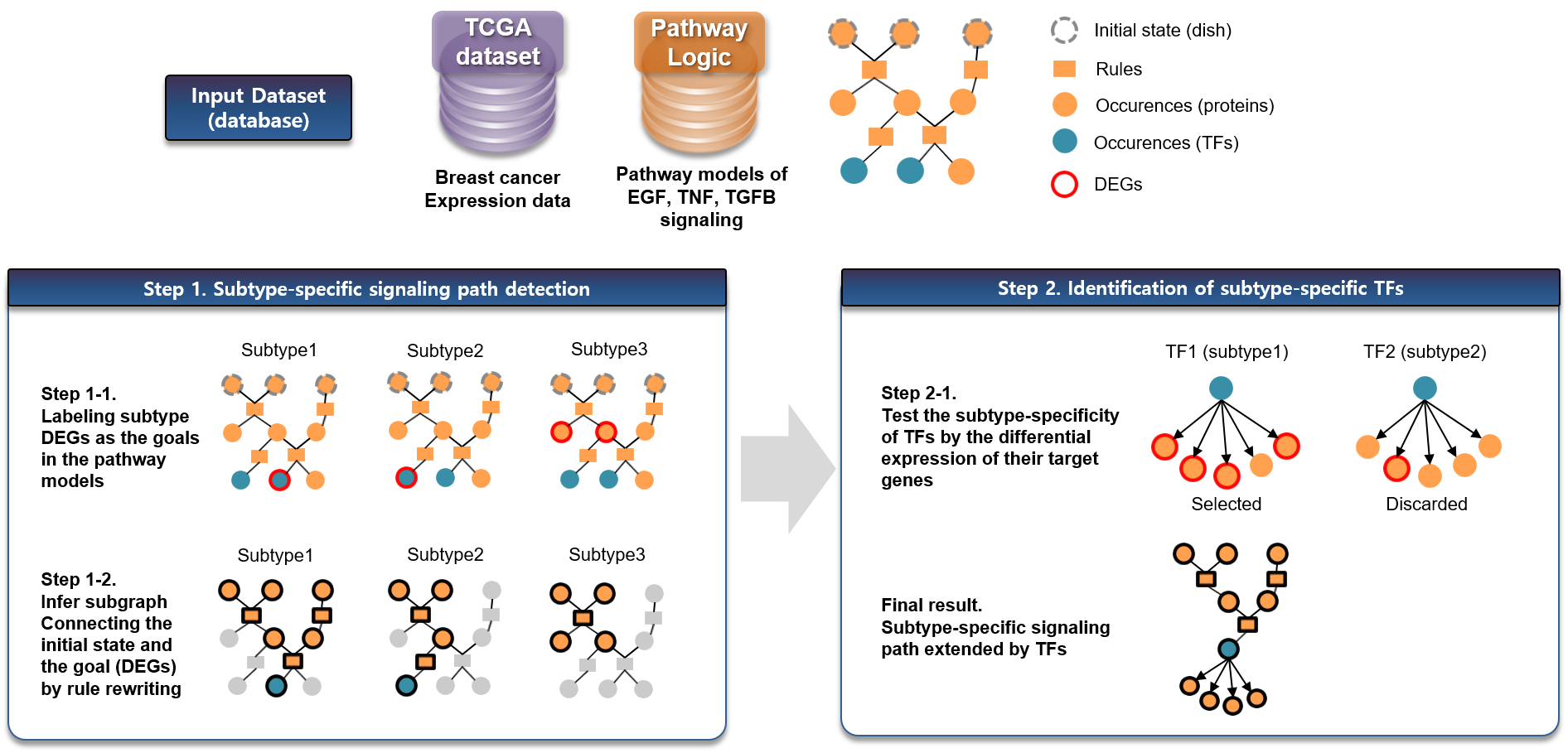
|
Pathway Logic is a rewriting-logic-based formal system for modeling biological pathways including post-translational modifications. Integrating gene expression data on PL system, we constructed subtype-specific path from key receptors (TNFR, TGFBR1 and EGFR) to key transcription factor (TF) regulators (RELA, ATF2, SMAD3 and ELK1) and to downstream genes targeted by TF. Proposed method identified potential pathway crosstalk via TFs in basal-specific paths, which could provide a novel insight on aggressive breast cancer subtypes. |
TimeTP
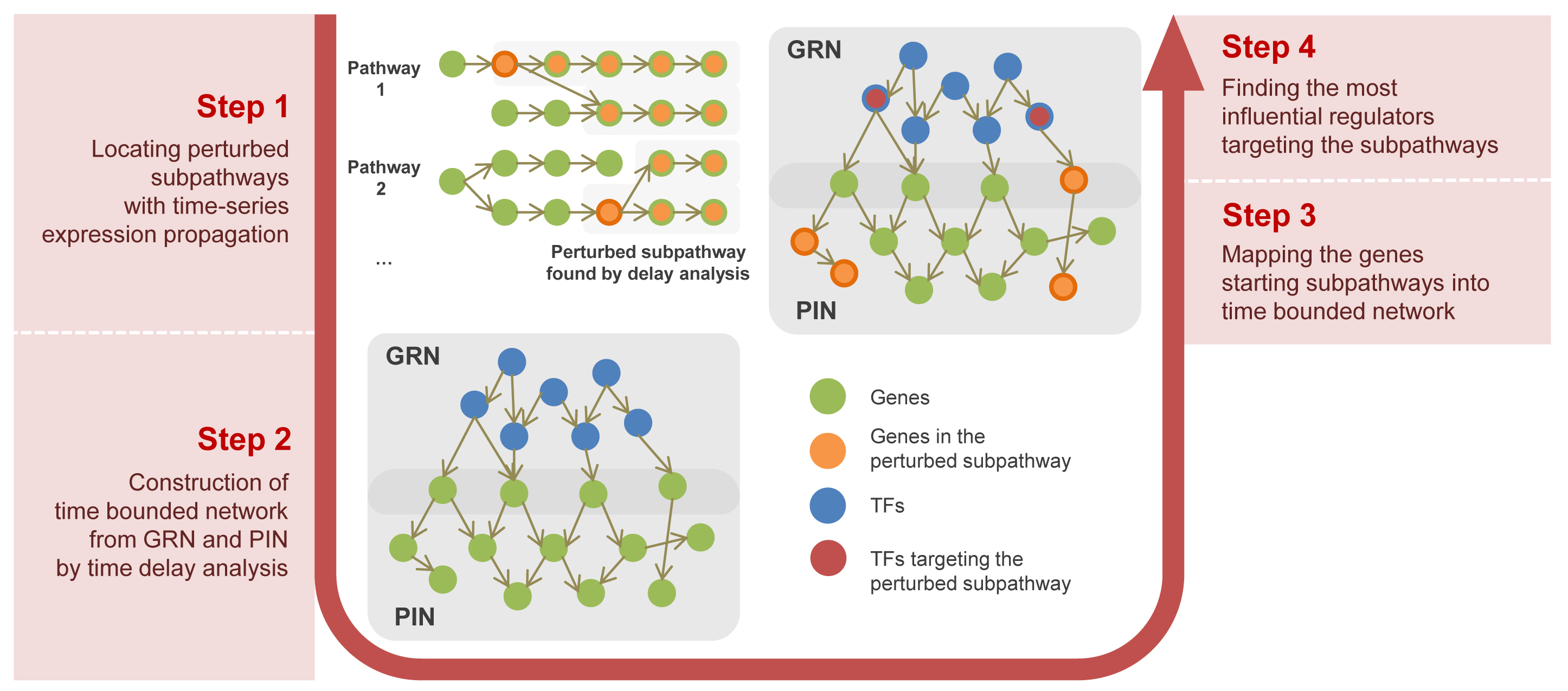
|
TimeTP finds perturbed sub-pathways and their regulators (transcription factors) from time series gene expression data. TimeTP uses cross-correlation to filter gene-gene pairs with similar expression patterns and locate perturbed subpathways. TimeTP performs the labeled influence maximization to find transcription factors that trigger the pathway perturbation. TF-Pathway map in time clock is generated to visualize path from TF to perturbed subpathway with expression propagation along time. |
TRAP
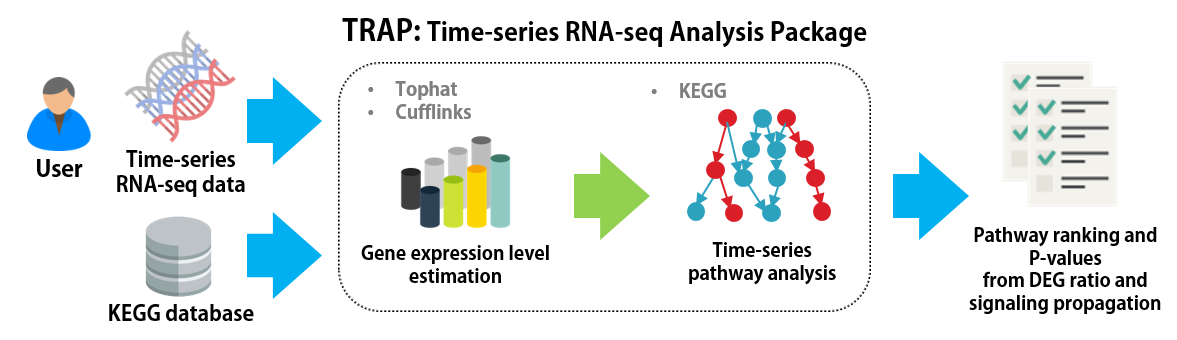
|
TRAP is a pathway analysis tool for time series gene expression data. TRAP estimates statistical values for each pathway that indicates the amount of activation/inhibition, considering the dynamic signaling propagation among genes. |